|
Bacterial displayBacterial display (or bacteria display or bacterial surface display) is a protein engineering technique used for in vitro protein evolution. Libraries of polypeptides displayed on the surface of bacteria can be screened using flow cytometry or iterative selection procedures (biopanning). This protein engineering technique allows us to link the function of a protein with the gene that encodes it. Bacterial display can be used to find target proteins with desired properties and can be used to make affinity ligands which are cell-specific. This system can be used in many applications including the creation of novel vaccines, the identification of enzyme substrates and finding the affinity of a ligand for its target protein. Bacterial display is often coupled with magnetic-activated cell sorting (MACS) or fluorescence-activated cell sorting (FACS) techniques. Competing methods for protein evolution in vitro are phage display, ribosome display, yeast display, and mRNA display. Bacteriophage display is the most common type of display system used [1] although bacterial display is becoming increasingly popular as technical challenges are overcome. Bacterial display combined with FACS also has the advantage that it is a real-time technique. HistoryCell display systems were first used in 1985, when peptides were genetically fused with proteins displayed on the M13 bacteriophage. Bacteriophage display is a commonly used cell display system, although it carries limitations in the size of proteins that can be displayed. Bacterial display was then introduced in 1986, allowing the surface display of larger proteins. Bacterial display systems were first introduced by Freudl et al. and Charbit et al. in 1986, when they used bacterial surface proteins OmpA and LamB to display peptides. Freudl et al. fused peptides with linkers with the ompA gene, causing the peptides to be expressed in the OmpA proteins. They showed that the proteins were now subject to cleavage by proteinase K. The non-OmpA peptides inserted were therefore a target of proteinase K. Insertion of the foreign peptides did not affect bacterial cell growth. Charbit et al. firstly defined the areas of the LamB protein that were "permissive" for foreign petide insertion (ie that did not lead to a complete loss of functionality of the protein). Then, they explored the versatility of the permissive sites (size limit, nature of the epitope,...) that were all located in surface-exposed loops of the trimeric outer membrane porin, aiming at developing multivalent live bacterial vaccines (12-15). This was the first evidence of using bacterial surface display techniques to express proteins on the surface of cells, without altering the function of the cell.[2] PrinciplePeptides are very useful as therapeutic and diagnostic substances. Their use is getting more popular, and display systems offer a useful way to engineer peptides and optimise their binding capabilities. Cells express surface proteins which can be involved in a whole host of responses including recognition of other cells, interaction with other cells, and cell signalling. Many types of bacteria have cell surface proteins such as the enteropathogenic E. coli intimin protein which is involved in binding to host cells, or the OmpA protein of E. coli cells which is important in keeping the structure of the outer membrane.[3] Many surface proteins are involved in bacterial cell attachment and invasion of the host cell. By using bacterial display, target proteins on the host cell can be identified. These surface proteins need to first be translocated across the bacterial cell membranes from the cytoplasm to the cell surface. Gram-negative bacteria have an additional periplasmic space, which Gram-positive bacteria lack, so they have a harder task of translocating proteins. The display of heterologous proteins on the bacterial cell surface normally requires the fusion of the protein with a surface protein, called a scaffold. 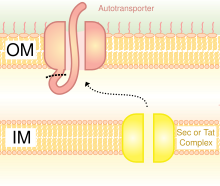 ScaffoldScaffolds are used to display the heterologous protein on the bacterial cell surface. There are various scaffolds which have been used such as outer membrane proteins, fimbriae/flagella proteins and CPX (circularly permuted OmpX).[4] The CPX scaffold allows peptide fusion at both termini of the scaffold. OMPs are common scaffolds for bacterial display. Proteins can also be displayed on the bacterial cell surface through the use of autotransporters. Autotransporters form part of the type V secretion system. They usually have three domains: leader sequence at the N-terminal; central passenger domain; autotransporter domain at the C-terminal. The heterologous protein is inserted at the passenger domain.[5] Another method of heterologous protein fusion is fusion with fimbriae/flagella, which are filamentous protrusions on the cell surface. There are many fimbriae on mainly Gram-negative bacteria, so displaying proteins on fimbriae is advantageous over some other surface proteins which are less numerous. A disadvantage of using fimbriae is that there is a relatively small insert size limit of 10-30 amino acids.[6]  Once the heterologous protein has been fused with the bacterial cell surface protein, it is exposed to either an enzyme, a cell (expressing a target protein) or an antibody (usually fluorescently tagged), depending on the application of the experiment. The sample is then passed through a beam of light during FACS, in a very narrow stream of fluid so that only one cell can pass at a time, and the fluorescence emitted is detected. Information on the size of the cell can be obtained by the scattering of light and if binding of the heterologous protein with the target protein/cell has occurred, there will be more fluorescence emitted. ApplicationsBacterial surface display can be used for a variety of applications. These include affinity-based screening, antibody epitope mapping, the identification of peptide substrates, the identification of cell-binding peptides and vaccine generation.[7] Affinity-based ScreeningScreening is used to find the apparent affinities of heterologous proteins displayed on the bacterial cell surface for target proteins. This method is usually combined with FACS, and the addition of a non-fluorescent target protein competitor is beneficial to obtaining more accurate binding affinities. Adding a competitor reduces the chance of target proteins rebinding, which would render the binding affinity less accurate. Cyclic peptide binders ScreeningCyclic peptides can be successfully displayed on bacterial cell surface.[8] By DNA randomization millions of cyclic peptides displayed on cell surface can be screened against a protein target using high-throughput FACS.[9] Antibody Epitope MappingAntibody epitope mapping is used to find the specificity of an antibody. The epitope (antibody binding site of antigens) is expressed on the bacterial cell surface by expressing a region of the gene encoding the antigen. Flow cytometry with fluorescently-labelled antibodies is used to detect the amount of antibody binding to epitope.[10] Identification of Peptide SubstratesThis can be applied to find the best substrates for proteolytic enzymes. The substrate is displayed on the bacterial cell surface between an affinity ligand and the scaffold, and the kinetics of substrate proteolysis is measured using FACS. Identification of Cell-binding PeptidesBacterial display can be used to find peptides which bind to specific cells e.g. breast cancer cells or stem cells. Displayed proteins are fluorescently tagged with GFP, so binding interactions between peptides and target cells can be seen by flow cytometry. Control samples are required in order to measure fluorescence levels in the absence of displayed peptides. Samples are also required which don’t contain displayed peptides, but contain mammalian cells and bacterial cells (including the scaffold). Vaccine DeliveryVaccine delivery is a very common application of bacterial surface display. There are two types of live bacterial vaccines that can be made:
Using bacterial surface display of antigens is a valuable alternative to conventional vaccine design for various reasons, one of them being that the proteins expressed on the bacterial cell surface can act favourably as an adjuvant. Conventional vaccines require the addition of adjuvants. Another advantage of generating vaccines using bacterial display systems is that the whole bacterial cell can be incorporated in the live vaccine [11] Unlike bacteriophage display systems which are generally used in vaccine development to find unknown epitopes, bacterial display systems are used to express known epitopes and the cells act as a vaccine delivery system.[12]
Comparison with phage displayUnder similar conditions, selection of bacterial-displayed peptides to model protein streptavidin proved worse.[13] References
12. Charbit A, Boulain JC, Ryter A, Hofnung M. Probing the topology of a bacterial membrane protein by genetic insertion of a foreign epitope; expression at the cell surface. EMBO J. 1986 Nov;5(11):3029-37. 13. Charbit A, Sobczak E, Michel ML, Molla A, Tiollais P, Hofnung M. Presentation of two epitopes of the preS2 region of hepatitis B virus on live recombinant bacteria. J Immunol. 1987 Sep 1;139(5):1658-64. 14. Charbit A, Molla A, Saurin W, Hofnung M.Versatility of a vector for expressing foreign polypeptides at the surface of gram-negative bacteria. Gene. 1988 Oct 15;70(1):181-9. 15. Newton SM, Klebba PE, Michel V, Hofnung M, Charbit A. Topology of the membrane protein LamB by epitope tagging and a comparison with the X-ray model. J Bacteriol. 1996 Jun;178(12):3447-56. |